Generic Marker of Cancer Prognosis
Our research has shown that disordered chromatin organization is an aberration found in multiple cancer types. Albeit the approaches differ slightly between the studies, it appears that the underlying principle is the same. Our vision is to unify the approaches to facilitate prognostication of patients based on their chromatin organization without having to learn specific attributes of each analyzed data set and each cancer type.
Texture analysis provides descriptions of chromatin structure in cell nuclei, however, the underlying biological mechanisms are poorly understood. We aim to improve our understanding of which mechanisms are involved to improve the prognostic ability of the marker and to reveal novel insight into carcinogenesis.
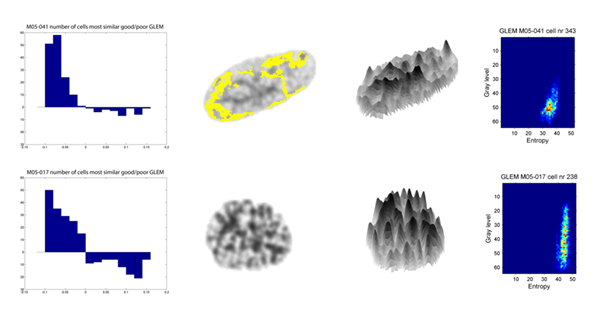
Texture features describe DNA density distributions in local regions in cancer cell nuclei, accumulated to a description of the chromatin structure on a nuclear level, as illustrated in the rightmost subfigure above. In prognostic studies we calculate texture features for millions of cell nuclei and identify the chromatin patterns that are most typical for patients with similar clinical outcome, e.g. death from disease. One approach in improving our understanding of chromatin organization is to map texture patterns back to the nuclei from which they originate and to study the spatial conservation of the patterns, e.g. whether prognostic information is mainly found near the nuclear periphery. The yellow regions in the cell nucleus illustrated above represent this approach. We have observed that cell nuclei containing texture patterns that are typical for e.g. poor prognosis are found in most patients, but in different proportions (illustrated in the leftmost figure above).
The approach described in the previous section may reveal new knowledge, but will not allow pinpointing of relevant mechanisms on a molecular level. Co-registration of chromatin patterns, protein expression and methylation patterns on a cell-by cell basis will allow us to reach this goal. We aim to map information with pixel precision, i.e. to associate subcellular texture feature patterns with data on a molecular level. Novel methods for multiple iterations of immunohistochemical staining and restaining with different antigens are key to this ambitious project that requires the development of advanced methods to align the resulting image data and a robust statistical framework to interpret the results.
Linking chromatin patterns and biological mechanisms
Texture analysis provides descriptions of chromatin structure in cell nuclei, however, the underlying biological mechanisms are poorly understood. We aim to improve our understanding of which mechanisms are involved to improve the prognostic ability of the marker and to reveal novel insight into carcinogenesis.
Texture features describe DNA density distributions in local regions in cancer cell nuclei, accumulated to a description of the chromatin structure on a nuclear level, as illustrated in the rightmost subfigure above. In prognostic studies we calculate texture features for millions of cell nuclei and identify the chromatin patterns that are most typical for patients with similar clinical outcome, e.g. death from disease. One approach in improving our understanding of chromatin organization is to map texture patterns back to the nuclei from which they originate and to study the spatial conservation of the patterns, e.g. whether prognostic information is mainly found near the nuclear periphery. The yellow regions in the cell nucleus illustrated above represent this approach. We have observed that cell nuclei containing texture patterns that are typical for e.g. poor prognosis are found in most patients, but in different proportions (illustrated in the leftmost figure above).
The approach described in the previous section may reveal new knowledge, but will not allow pinpointing of relevant mechanisms on a molecular level. Co-registration of chromatin patterns, protein expression and methylation patterns on a cell-by cell basis will allow us to reach this goal. We aim to map information with pixel precision, i.e. to associate subcellular texture feature patterns with data on a molecular level. Novel methods for multiple iterations of immunohistochemical staining and restaining with different antigens are key to this ambitious project that requires the development of advanced methods to align the resulting image data and a robust statistical framework to interpret the results.
This text was last modified: 14.08.2017